Radical ions, which consist of unpaired electrons, offer distinct gas-phase ion chemistry as compared to the even-electron species. Radical chemistry can be utilized to tackle challenging problems, such as differentiating isomeric structures, which would otherwise not be solved by traditional MS analysis of even-electron ions of the biomolecules. We are developing MS instrumentation and methods to facilitate radical reactions for either in the vacuum or in ambient air.
1. Radical reactions at the interface of ESI-MS.
Radicals or excited neutrals are generated via air discharge or UV photolysis and subsequently reacted with ions entrained in the ESI plume. Radical reactions are subsequently monitored and characterized in situ by MS analysis. Reactions of peptides and lipids with various radical species have been investigated, including •OH, •CH2OH, excited state of (CH3)2CO. Novel analytical applications based on these reactions have been developed.
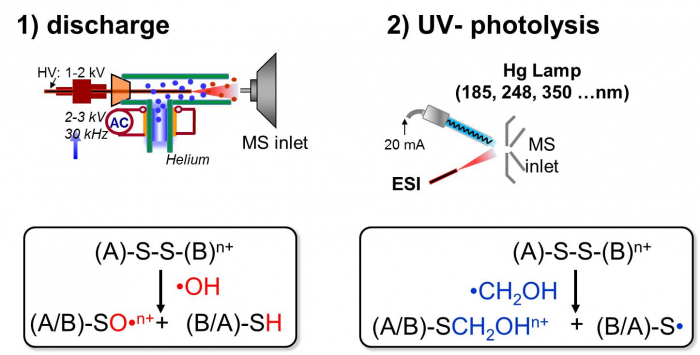
AC; 2010, 82, 2856; JASMS., 2011, 22, 922; Analyst, 2013, 138, 2840;JASMS, 2014, 25, 1192.
2. Ion/radical reactions in a linear ion trap mass spectrometer.
A first linear ion trap mass spectrometer capable of studying reactions between the mass-selected ions and radicals has been recently developed and tested in collaboration with Prof. Zheng Ouyang from Biomedical Engineering at Purdue. This instrument uses a rectilinear ion trap as the mass analyzer and gas-phase reactor, an ESI as the source of the bimolecular ions, a pulsed pyrolysis valve for the generating an intense radical beam, and a glow discharge electron impact (GDEI) source for radical characterization. This MS platform can facilitate mechanistic studies on the radical attack to biomolecules that are of biological significance
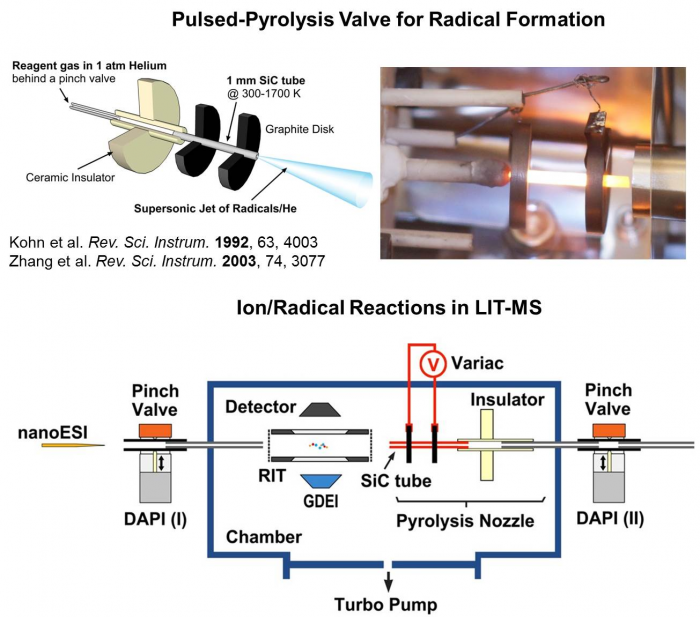
3. Chemistry of bio-radical ions.
Through radical relations at ESI-MS interface, our group has synthesized and studied cysteine sulfinyl radical in the gas phase (Cys-SO•), which has a wide relevance to radical-induced oxidation of proteins, however, has been poorly characterized due to its transient nature in the condensed phases. Different from carbon-centered radicals, we have discovered that sulfinyl radical has a dual property of being acting as a base or a radical via combined experimental and theoretical approaches. The base property allows the formation of proton bridging between the radical site and the neighboring amino acid residues and thus contributes to the overall structural and chemical property of a polypeptide.
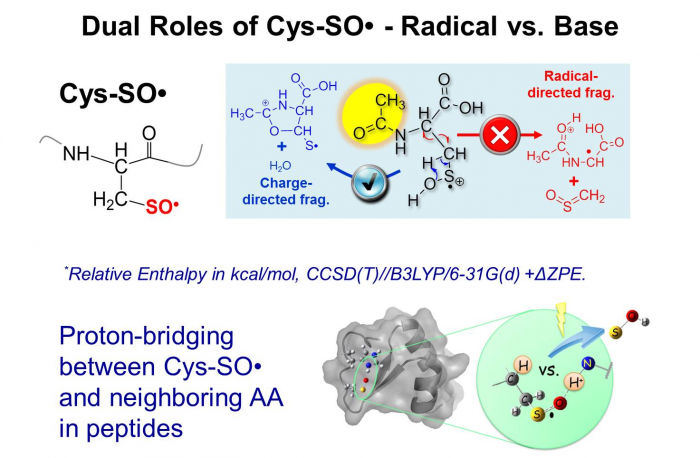
Love et al. J. Am. Chem. Soc., 2013, 135, 6226
Tan et al. J Phys. Chem. A, 2014, 118 ,11828
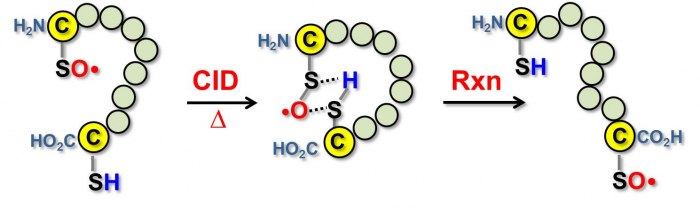
We also systematically investigated the inter- and intra-molecular reactivity of the sulfinyl radicals. They showed that the cysteine sulfinyl radical can react with a disulfide bond or a thiol group within a peptide, which has implications to radical-induced disulfide bond scrambling.
Using functionalized sulfinyl radical as a precursor of glycyl-type radical, we have also developed an experimental approach based on tandem mass spectrometry to correlate the electronic property of the connecting groups to the stability of glycyl-type radical species (Angew. Chem., Int. Ed., 2014, 53, 1887-1890, featured as the front cover and the “hot article”).
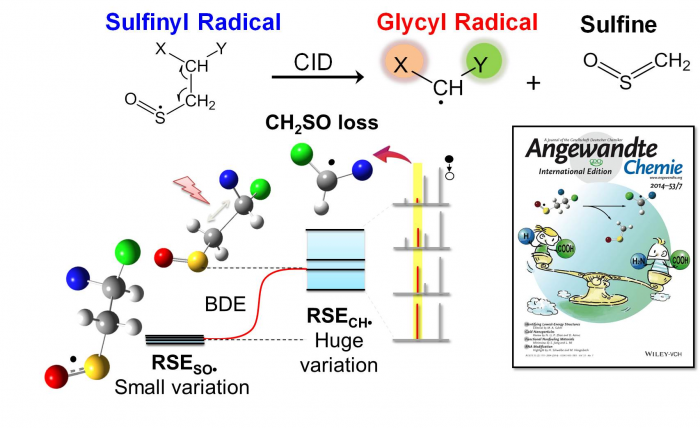
Tan et al. Angew. Chem. Int. Ed. 2014, 53, 1887
4. Application of radical chemistry for bio-analysis Analysis of unsaturated lipids:
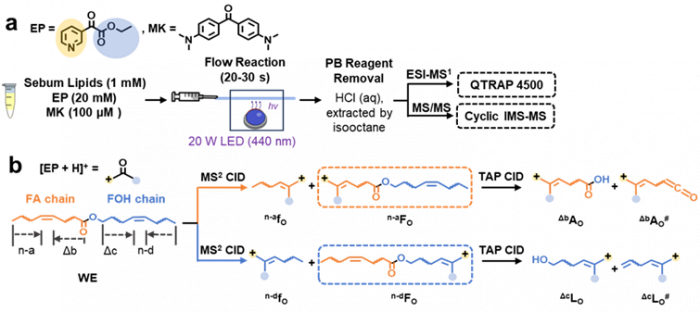
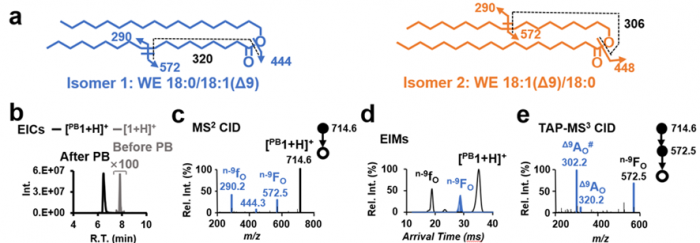
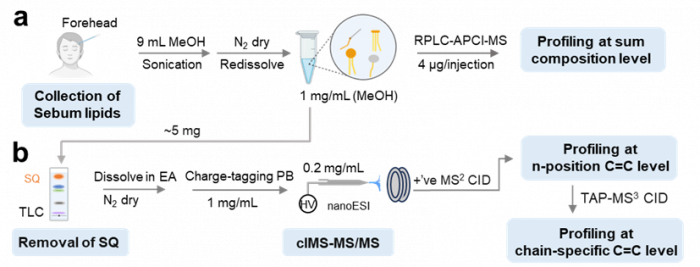
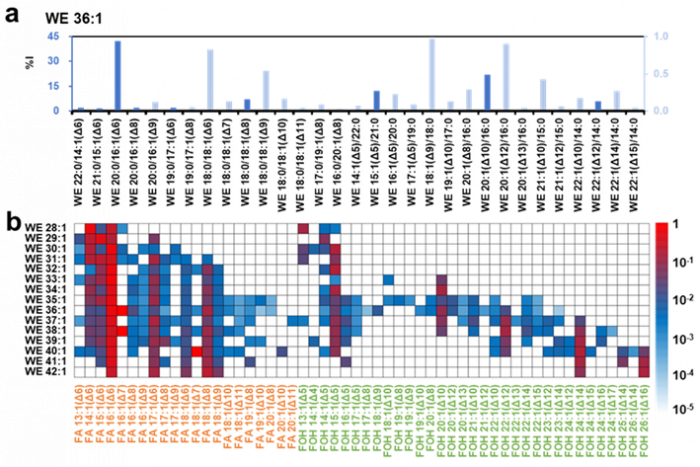
Facile determination of C=C bond locations of lipids is a long-standing challenge for lipid analysis using MS. Intact lipid analysis via conventional low energy collisional activation tandem mass spectrometry does not provide information for the C=C location because much higher energies are required for cleaving C-C or C=C bonds and thus no fragments specific to the C=C locations can be produced. Utilizing the high reactivity of C=C with radicals or electrophilic excited state molecules, our group has recently developed coupling Paternò–Büchi (PB) reaction with MS/MS for highly confident C=C bond location determination in lipids (Angew. Chem., Int. Ed, 2014, 53, 2592-2596). This PB-MS/MS strategy is currently being developed for unsaturated lipid C=C location isomer characterization and quantitation of biological samples (tissue, cell lines, plasma), application to shotgun and separation based lipidomics, biomarker discovery, and bio-imaging.
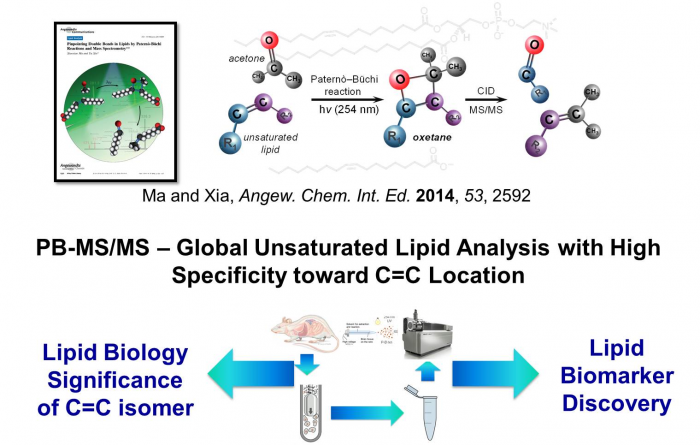
Ma and Xia, Angew. Chem. Int. Ed. 2014, 53, 2592