News & Views | Published: 05 February 2015
Molecular histology
More than a picture
Richard W. Vachet
Department of Chemistry, University of Massachusetts Amherst, Amherst, Massachusetts 01003, USA
Nature Nanotechnology, 2015, 10, 103–104
A label-free mass spectrometry imaging method maps the locations of carbon nanomaterials injected into mice through the detection of small carbon clusters.
The old saying that 'a picture is worth a thousand words' is no less true in science. Our understanding of biological processes in different organs has long been informed by pictures from histological studies. Recently, there has been tremendous interest in adding a molecular component to such images. Molecular histology, as it is often called, can reveal the underlying biochemistry of tissues and organs, while simultaneously providing information on how therapeutics or toxins influence the function or misfunction of an organ. Now, writing in Nature Nanotechnology, Zhongxiu Nie and colleagues at the Chinese Academy of Sciences and Ohio State University describe a mass spectrometry-based method to perform molecular histology on carbon nanomaterials in tissues of mice1. The imaging approach offers a powerful way to quantitatively evaluate the fate of carbon nanomaterials in vivo.
Imaging mass spectrometry has emerged as a unique tool by which individual molecular species can be directly detected in tissues with spatial resolutions around 20 μm or less2. The technique is inherently 'label free' because all molecules have mass, and as long as they can be ionized, they can be detected. One of the most common methods for ionizing molecules in imaging mass spectrometry is by matrix-assisted laser desorption/ionization (MALDI), whereby an organic matrix (usually in an excess of 10,000 to 1 molar ratio over the analytes) is typically co-crystallized with a sample of interest and then irradiated with a laser3. The matrix absorbs the laser irradiation, is rapidly excited and vapourized, and subsequently the co-crystallized analytes are transferred into the gas phase, where the analyte can be ionized by charge transfer from the matrix. However, no one has demonstrated the ability of MALDI to detect intact carbon nanomaterials because it is challenging to co-crystallize appropriate matrices with carbon nanomaterials. Without a matrix, it is difficult to desorb the analytes intact into the gas phase.
To solve this problem, Nie and co-workers forgo the use of the traditional matrix and rely on the strong absorbance of carbon nanomaterials in the near-ultraviolet region of the electromagnetic spectrum. Because most biological molecules do not absorb light very well in the near-ultraviolet, the result is the selective detection of small carbon clusters (C2–C10) arising from the desorption and ionization of carbon nanomaterial fragments with almost no background signal from any biologically derived molecules. By rastering the laser across tissue slices, individual mass spectra can be obtained, and the carbon cluster signals can then be used to generate an image that shows the locations of the carbon nanomaterial (Fig. 1). When used with appropriate standards, the quantities of the carbon nanomaterials at given locations can be determined.
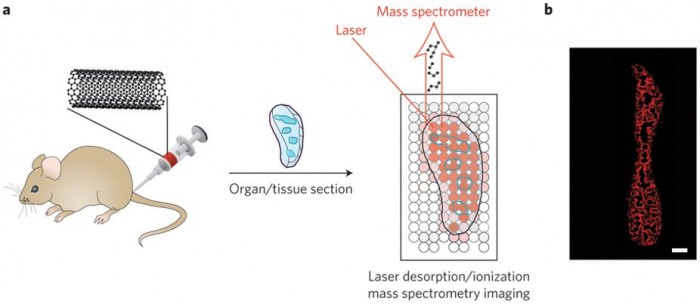
Figure 1: Laser desorption/ionization mass spectrometry imaging of carbon nanomaterials in tissues. a, Carbon nanotubes, graphene oxide, or carbon nanodots are injected into mice. The mice are killed, organs of interest are taken and sectioned, and laser desorption/ionization mass spectrometry is used to image the locations of the carbon nanomaterials (blue outlines). The laser is rastered across the tissue, and mass spectra are acquired at each spot. b, Images are generated by plotting carbon cluster ion intensities as a function of location in the tissue. Shown here as an example is an image of a spleen from a mouse treated with carbon nanotubes. Red areas are where carbon nanotubes are found. Panel b reproduced from ref. 1, 2015 Nature Publishing Group.
There are several merits to this mass spectrometry-based molecular imaging approach. Because it is label free, the carbon nanomaterials do not need to be modified for them to be detectable. Furthermore, there is no need to worry about any loss of label in vivo, or to account for the way the label might affect the biological distributions of the nanomaterials. Moreover, Nie and co-workers showed that the method can provide quantitative information about carbon nanotubes, graphene oxide and carbon nanodots in a variety of tissues. The quantitative images reveal valuable insights into how the carbon nanomaterials are trafficked and processed by different organs. For example, images of the spleen clearly show that carbon nanomaterials concentrate in the marginal zone between the red and white pulp. Because the marginal zone is responsible for presenting circulating antigens to lymphocytes (a type of white blood cell) in the spleen, this suggests an immune response4. Such information is difficult to obtain by any other means.
Another key advantage of the approach is that it should, in principle, work for other types of nanomaterial that absorb well in the near-ultraviolet. Nie and colleagues demonstrate that molybdenum disulfide nanosheets can also be monitored, and a recent study has shown gold nanoparticles can be imaged using a similar approach5.
As promising as the method is, detecting carbon cluster ions as proxies for the carbon nanomaterials means that the method does not directly indicate whether the carbon nanomaterials have been transformed in vivo. Any reactions (for example oxidation) undergone by the nanomaterials might be apparent in any new cluster ions that appear, but knowing the extent to which the nanomaterials are degraded would seem beyond the capabilities of the current imaging method.
Nonetheless, imaging mass spectrometry has a bright future when it comes to studying nanomaterials in vivo. One particularly exciting advance would be to extend the reported approach to enable the simultaneous detection of nanomaterials and nearby proteins or other biomolecules. Images that reveal what biological molecules orient themselves around nanomaterials would provide an even deeper understanding of the biochemical effects of such materials. One would think that pictures such as this might then be worth ten thousand words or more.
https://www.nature.com/articles/nnano.2015.4
References
- Chen, S. et al. Nature Nanotech. 10, 176–182 (2015).
- Chughtai, K. & Heeren, R. A. Chem. Rev. 110, 3257–3277 (2010).
- Norris, J. L. & Caprioli, R. M. Chem. Rev. 113, 2309–2342 (2013).
- Mebius, R. E. & Kraal, G. Nature Rev. Immunol. 5, 606–616 (2005).
- Yan, B. et al. J. Am. Chem. Soc. 135, 12564–12567 (2013).




